Catherine Ott
Spring 2001
Colony
Dynamics
Bacteria grown on
agar surfaces can grow into a variety of colony shapes, ranging from a basic
circle to an intricate fractal-like pattern. The diversity is caused by the
motility of the bacteria, which is the result of the affects of agar
concentration and nutrient concentration, as well as numerous other factors.
Many biologists, physicists, and mathematicians have studied the colony
formations. Each group of scientists attempts to develop a model that will
represent all of the possible formations. The study of these models, as well as
observing bacteria and performing experiments, has lead to further understanding
of the systems influencing the final shape of the colony. The observation of
bacteria moving laterally (which is not a direction swimming bacteria can move)
has prompted questions about the fluid dynamics of the colonies. Fluid-like
motion has been observed within the colonies, and the origin of this motion must
be determined for proper incorporation into the models. An experiment has been
performed to make the conclusion that solely the motion of the bacteria
themselves causes the fluid flow; further experiments are being developed.
Motile strains of
bacteria, such as Bacillus subtilis, swim by using flagella, which are
situated around their oblong bodies. The flagella propel the cells forward only.
When cells are placed on an agar surface of suitable wetness, the bacteria
reproduce and form colonies of swimming cells. Through chemotaxis, oriented
movement toward or away from a chemical stimulus, the cells find regions of
higher nutrient concentration. Also, the cells diffuse across the agar.
Diffusion is the spreading of the bacteria, or any material, down their own
concentration gradient. These motions of bacteria result in diverse colony
shapes, depending on parameters such as agar concentration and nutrient
concentration. In softer agar, the bacteria can swim and diffuse easily. In
fact, in very soft agar the bacteria actually swim inside the agar matrix.
Nutrient concentration affects the strength of the cells, and their resulting
ability to swim and reproduce. Scientists have tried many techniques to develop
a model that accounts for diffusion and chemotaxis, and can generate each of the
following forms under different initial conditions.

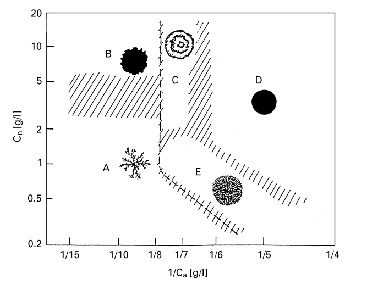
Figure 1.
Morphological diagram of B. subtilis colony patterns. From Mimura, et al.
[6]
A) diffusion-limited
aggregation-like (DLA);
B) Eden-like;
C) concentric
ring-like;
D) disk-like;
E) dense branching
morphology-like.
The goals of my work
are to understand models developed by other scientists, observe and experiment
with bacteria cultures, and contribute ideas toward the development of new
models.
One of the first
models of bacteria movement was developed by Evelyn Keller and Lee Segel, as
reported in their paper "Traveling Bands of Chemotactic Bacteria: A
Theoretical Analysis." [3] Keller and Segel used partial differential
equations to model bacteria swimming in a tube. The bacteria traveled in bands
toward nutrients and oxygen, coined "critical substance." The model
expressed the motion of bacteria through chemotaxis and diffusion, but growth
was not considered. Also, the motility of the bacteria was assumed to vary with
the substrate concentration, and not with the bacteria concentration. The
critical substance does not diffuse a significant amount, and there is enough
substance such that availability is not a limiting factor. The bacterial flux
due to chemotaxis is proportional to the critical substance gradient. Traveling
wave solutions of this system can be obtained. The derivation of the Keller-Segel
model can be found in Michelle Cobeaga's report "Analysis of Migrating
Bands of Chemotactic Bacteria." [1]
Another factor that
some scientists feel is important is the excretion of a lubricating fluid by the
bacteria. The reason that this could be important is that the degree of wetness
of the agar is a limiting factor for the growth of a branch. In very dry agar,
the expansion of the colonies is not related to motility of the bacteria; the
colony spreads as a consequence of the bacteria growing and demanding more area.
The secretion of a fluid would make a path of less resistance, and more cells
would follow. Mendelson's "Pattern of Reporter Gene Expression in the
Phase Diagram of Bacillus subtilis Colony Forms" [5] demonstrates
that the breaking of a boundary of a branch is achieved by an accumulation of
cells, as the surface tension of the water in the agar is too high for
individual cells to overcome. So, a branch could be created by groups of cells
lubricating the agar and allowing more cells to follow this path. Golding et al.
reported on this in "Studies of Bacterial Branching Growth using
Reaction-Diffusion Models for Colonial Development." [2] Also, the velocity
of the bacteria was used to define the different colony shapes. Four of the
patterns, DLA patterns, compact patterns, and dense branches and concentric
rings, corresponded to three different velocities. This means that the patterns
take different amounts of time to develop. Their conclusion was that the
morphologies are the result of actual transitions in the way the bacteria move
and the velocity of movement. One issue that Golding et al. pointed out is that
two different mathematical descriptions can lead to patterns that appear
similar. An example of this would be obtaining branches by including a bacteria
death term, and also by having no death term but assuming the movement of the
bacteria is dependent on the density of bacteria and the density of food. These
are distinctly different models, but simulations show similar results.
A paper by Mimura et
al., entitled "Reaction-Diffusion Modeling of Bacterial Colony
Patterns" [6] begins by asking a question: "Is the diversity of such
colony patterns caused by different effects or governed by the same underlying
principles?" To answer this question, the authors first critiqued a variety
of models developed by other scientists. The models are based on
reaction-diffusion (RD) concepts, in which "spatial and temporal change of
bacteria are described by using their average densities." The models
discussed included Kessler and Levine's cutoff mechanism in the growth term,
which allowed the development of branching patterns. Kawasaki et al. assumed
that the diffusion coefficient of bacteria depends on their own density, the
density of the nutrients, and a stochastic term to introduce frontal
instability. These ideas led to a nonlinear diffusion model. Finally,
Kitsunezaki developed a density-dependent model with an interesting term that
separated bacteria into groups with different motility properties. The bacteria
are divided into those that are active, and bacteria that are inactive; this
idea is very similar to Golding et al.'s death term. This model also produces
frontal instability that results in branching patterns. Mimura et al. proposed
an improved model, which accounts for the motility rate of the active bacteria,
the diffusion rate of nutrients, the growth rate of the bacteria, and the rate
of conversion of active bacteria into inactive bacteria. The primary difference
from the previous models is in the death term. Mimura et al. use a piecewise
function that is dependent on the bacterial and nutrient concentrations to model
the death of bacteria. This model can produce four different types of patterns
by changing the motility of bacteria (dependent on the concentration of agar)
and the nutrient concentration. The Eden-like patterns (shown in Figure 1.1) can
be reproduced from a proposed nonlinear diffusion model.
While these models
have shown an accurate description of the end result of a colony shape, the
process of achieving that shape is much more detailed. Cells have three main
levels of organization, as described by Mendelson, et al. in "Organized
Cell Swimming Motions in Bacillus subtilis Colonies: Patterns of
Short-Lived Whirls and Jets." [4] The first level is that of individual
cell motion. Secondly, groups of cells form whirls and jets. What is most
interesting is that after an opposing jet disorganizes a whirl, the whirl
reorganizes into a whirl in the opposite direction of what it started. To show
that this is actually a function of bacteria motion, marker beads were added to
the colony’s advancing finger. While the markers did participate in whirls and
jets, they did not reverse direction not retrace an earlier route. The third
level of organization occurs when the whirls and jets organize into a
superpattern. This superpattern withstands the individual pattern elements
reorganizing. Ultimately, this interconnectedness is what forms extensive colony
morphology. When the whirls and jets strike a boundary, some cells are left at
the edge and push it outward; this is colony expansion. What should be
determined is what causes the swimming patterns. Could it be that the path is
the result of the physics of swimming, or is this chemotactic response towards a
self-emitted attractant?
The bacteria
strains, M8, M22(81), and M22(84), were obtained from Mendelson. They were
maintained on standard tryptose blood agar base (TBAB) plates made from Difco
medium. For growing colonies of the morphologies described above, a softer
version was made, which contained only 0.6% agar, instead of 1.5%. The method
for making this media [4] is to dissolve 10 grams (g) of tryptose, 3 g of beef
extract, and 5 g of NaCl in 1.0 liters of deionized water. Six grams of agar was
added. To sterilize, the mixture was autoclaved at 121°
C for 20 minutes. The solution was cooled to 48° C and maintained at this temperature for one hour. Then, the plates
were poured and the agar was allowed to solidify at 23°
C in a 50% relative humidity chamber. For clearer resolution on a microscope,
this media could be melted and dropped onto a cover slip. Then, to keep the
media from drying out, the cover slips were placed on a specimen slide, and this
was set on a bent glass rod, in a petri dish with a small amount of water. These
plates were stored in an incubator at 24° C. For viewing, the cover slip would be taken off the slide and placed
under the microscope. This ensures a minimum amount of glass and agar between
the microscope and the bacteria. Cultures were inoculated using sterile
toothpicks. The colonies were viewed with a Nikon-inverted phase-contrast
microscope with a 20x or 40x objective. Using a Cohu camera fitted directly to
the microscope, the images were transferred to VHS tape on a JVC or GYYR tape
deck, and recorded on a standard cassette. Then, the films were transferred into
a Dell computer and the images were analyzed using Matrox Inspector, Image-Pro
Plus, and Adobe Photoshop. Also, some frames were printed onto transparencies
using a Tektronix Phaser II SDX. An experiment using formaldehyde to kill the
bacteria was performed. In this experiment, cultures were grown on cover slips
in the standard way. Then, the cover slips were placed under the microscope with
a petri dish lid over them. A strip of paper towel wet with water was taped to
the inside of the lid, to keep the cultures moist and active. Another lid was
set up in the same way, but the paper towel was moistened with formaldehyde
instead of water. Fumes were allowed to build up inside the lid, and then the
lids were switched. The bacteria were almost immediately engulfed in
formaldehyde fumes.
The observations and
experiments serve to reinforce the previous observations described by Mendelson
et al., and also lead to a new observation. I saw the whirls and jets I had read
about, and noticed the patterns of disorganization and reorganization. By
varying the agar concentration, I witnessed the bacteria trapped within the
agar, bouncing between the walls of the pockets. I was also able to capture
clear images of the whirls depositing cells at the edge of the colony, and the
colony expansion that results. Performing the formaldehyde experiment described
above, as devised by Mendelson, showed the motion of the bacteria ceased within
seconds of introducing the formaldehyde into the system. The most exciting
observation, noticed by Mendelson, is of the parallel alignment of cells while
in whirls and jets, in which groups of cells are carried perpendicular to the
direction of movement cells are capable of through their own swimming. This
lateral motion, and resulting compression of the bacteria, is the result of a
force other than that of the bacteria exhibiting the motion. So, either the
bacteria create a fluid flow which pushes the bacteria, or there is an external
force creating the flow.
Most of the
observations and experiments only served to reinforce previous knowledge, and
give me first hand experience with the techniques and observations made by
others. I witnessed the advancement of the colony edge by depositing cells, and
how the wetness of the agar is a factor in growth. The rate of edge growth can
be compared to the rate of movement of the individual cells caught inside the
agar. This will give an indication of affect of the agar concentration on
movement. The formaldehyde experiment proves that only the force of the bacteria
causes the flow of the fluid and the resulting lateral movement of the groups of
cells; one group of cells is able to push another group of cells. Also, the rate
of the compression of the cells is in the process of being analyzed. These ideas
will increase the understanding of the dynamics of colony growth.
Reading the
discussions of various models and observing the colonies myself has led me to a
few conclusions about the current models. Golding et al. [2] expressed that
including terms for "diffusion, food consumption, reproduction, and
inactivation" do not sufficiently describe the phenomena, and they propose
to include chemotactic signaling. While this is an improvement, I do not feel
that this will adequately describe the dynamics within the colonies. The models
I have read about focus on what happens at the edges to determine the colony
shape. My observations of the colonies have shown me that the rest of the colony
is important as well. Mendelson et al. [4] describe the levels of organization
of bacteria movement and resulting colony development, and I feel that the
interconnectedness of the movements can play an important role in developing a
comprehensive model.
1.
Cobeaga, Michelle. 2001. Analysis of Migrating Bands of Chemotactic
Bacteria.
2.
Golding, I., Y. Kozlovsky, I. Cohen, and E. Ben-Jacobs. 1998. Studies of
bacterial branching growth using reaction-diffusion models for colonial
development. Physica A. 260: 510-554.
3.
Keller, E. F., and L. A. Segel. 1971. Traveling bands of chemotactic
bacteria: a theoretical analysis. J. theor. Biol. 30: 235-248.
4.
Mendelson, N. H., A. Bourque, K. Wilkening, K. R. Anderson, and J. C.
Watkins. 1999. Organized cell swimming motions in Bacillus subtilis colonies:
patterns of short-lived whirls and jets. J. Bacteiol. 181: 600-609.
5.
Mendelson, N. H., and B. Salhi. 1996. Patterns of reporter gene
expression in the phase diagram of Bacillus subtilis colony forms. J.
Bacteriol. 178: 1980-1989.
6.
Mimura, M., H. Sakaguchi, and M. Matsushita. 2000. Reaction-diffusion
modelling of bacterial colony patterns. Physica A. 282: 283-303.